Transcriptomics: an introduction (lecture)
This presentation introduces transcriptomics, a field of biology focused on the comprehensive analysis of gene expression. It details the experimental process, from sample preparation and biomaterial isolation to RNA quantification and transcript measurement using microarrays and RNAseq. The presentation then explains the basic steps required for data processing, statistical analysis (including differential expression analysis and FDR calculation), and functional interpretation using techniques such as enrichment analysis, using by functional classifications such as gene ontology and pathway, and how this data can be used to support network analysis. Finally, it addresses data management, repositories (like GEO), and the importance of metadata to support the generation of robust and reliable results.
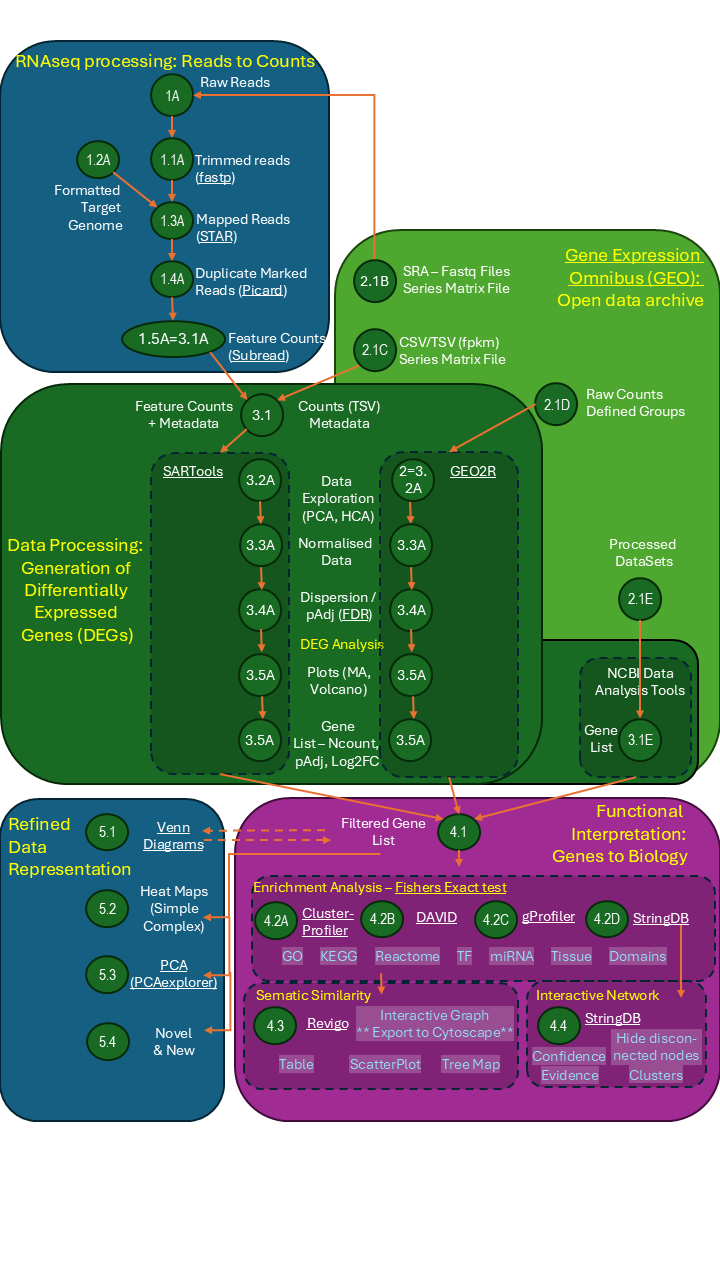
Transcriptomics Analytical Workflow
I have created a graphical overview of the data analytical workflow needed to analyse Transcriptomic data either by perfroming RNASeq analysis from your own raw sequence reads or through RNASeq or microarray data deposited in NCBI Gene Expression Omnibus (GEO) data repository. All research articles published in peer-reviewed journals must submit the data (raw or processed) to an accessible data archive - the most commonly used being NCBI GEO which allows you to reanalyse data that has been previously published and perform larger metadata studies using multiple data sources.
I have provided both a graphics version of the workflow but also a downloadable PDF which has live hyperlinks to software sources and instructions. Many new software packages are being developed and you should use these instructions as a framework adding new innovations as they become available.